 |
| Protein Post 3 |
Proteins
and Amino Acids:
Amino
acids always contain Carbon, Hydrogen, Oxygen and Nitrogen, and many in
addition carry Sulphur. In human Bio-Chemistry, 20 Amino acids are used as the
principal building blocks of protein, although there are other; for instance,
there are some amino-acids used only in certain proteins and some are seen only
in microbial products. Of The amino acids used in human protein synthesis,
there is a basic common structure, including an amino
group (NH2) a carboxyl
group (COOH) and a hydrogen atom. What makes one amino acid different from
next is a variable side chain. As in formation of glycosidic linkage, when two
amino acids join up the reaction expels a molecule of water and the resulting
bond is called a peptide bond.
Proteins
are made from amino acids joined together, and are the main family of molecules
from which the human body is built. Protein Molecules vary enormously in size,
shape, chemical constituents and function. Many important groups of
Biologically active substance are proteins:
1)
Carrier molecules e.g. Haemoglobin.
2)
Enzymes.
3) Many
hormones e.g. Insulin.
4)
Antibodies.
5) Motor
Protein
6)
Receptor Protein e.g Rhodopsin
7)
Structural Proteins
8)
Storage Proteins
etc.
Proteins
can also be used as an alternative energy source, usually in dietary
inadequancey, although the process in much less efficient than when
Carbohydrates or fats are broken down.
Primary
Structure of Proteins:
A
covalent bond forms between the amino group of one amino acid and the carboxyl
group to another. This covalent linkage, called a peptide bond, results in a
molecule called dipeptide. Three or more amino acids linked together form
polypeptide chain. Which kind of amino
acid follows another in the chain is always the same for all proteins of a
given type. For example, the two chains making up the protein Insulin always
have the sequences same. The specific sequence of amino acids in a polypeptide
chain constitute the primary structure of protein.
Secondery
Structure of Proteins:
The term
refers to the helical or extended pattern brought about by Hydrogen bonds at
regular intervals along a polypeptide chain. Some amino acids tend to favour
helical pattern, other tend to favour sheetlike patterns.
Tertiary
Structure of Proteins:
Protein
structure is also affected by interactions among R groups. Most helically
coiled chains become further folded into some characteristic shape when one R
group interacts with another R group some distance away, with the backbone
itself, or with other substances present in the cell. The term tertiary
structure refers to the folding that arises through interaction among R groups
of a polypeptide chain.
Quaternary
Structure of protein:
The 4th
level of protein in architecture, results from interactions between two or more
polypeptide chains in some protein. The resulting protein can be
globular,fiberlike,or some combination of the two shapes e.g. Haemoglobin.
 |
| Brief Pictorial Summery of Proteins |
Structure
determination
Discovering
the tertiary structure of a protein, or the quaternary structure of its
complexes, can provide important clues about how the protein performs its
function. Common experimental methods of structure determination include X-ray
crystallography and NMR spectroscopy, both of which can produce information at
atomic resolution. However, NMR experiments are able to provide information
from which a subset of distances between pairs of atoms can be estimated, and
the final possible conformations for a protein are determined by solving a
distance geometry problem. Dual polarisation interferometry is a quantitative
analytical method for measuring the overall protein conformation and
conformational changes due to interactions or other stimulus. Circular
dichroism is another laboratory technique for determining internal beta sheet/
helical composition of proteins. Cryoelectron microscopy is used to produce
lower-resolution structural information about very large protein complexes,
including assembled viruses; a variant known as electron crystallography can
also produce high-resolution information in some cases, especially for
two-dimensional crystals of membrane proteins. Solved structures are usually
deposited in the Protein Data Bank (PDB), a freely available resource from
which structural data about thousands of proteins can be obtained in the form
of Cartesian coordinates for each atom in the protein.
Many more
gene sequences are known than protein structures. Further, the set of solved
structures is biased toward proteins that can be easily subjected to the
conditions required in X-ray crystallography, one of the major structure
determination methods. In particular, globular proteins are comparatively easy
to crystallize in preparation for X-ray crystallography. Membrane proteins, by
contrast, are difficult to crystallize and are underrepresented in the PDB.
Structural genomics initiatives have attempted to remedy these deficiencies by
systematically solving representative structures of major fold classes. Protein
structure prediction methods attempt to provide a means of generating a plausible
structure for proteins whose structures have not been experimentally
determined.
Different Types of Proteins :
 |
| 1) Enzyme Protein |
 |
| 2) Gene Regulatory Protein |
 |
| 3) Motor Proteins |
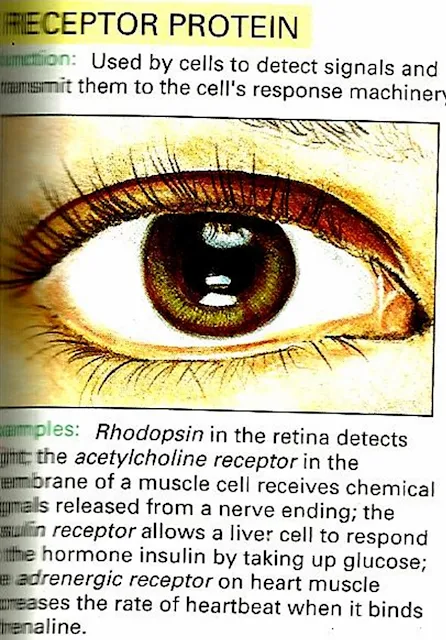 |
| 4) Receptor Protein |
 |
| 5) Signaling Proteins |
 |
| 6) Proteins For Special Purposes |
 |
| 7) Storage Proteins |
 |
| 8) Structural Protein |
 |
| 9) Transport Protein |
Nucleotides:
These are
the largest molecules in the body and are built from components called
nucleotides, which consists of three subunits are : A sugar ( ribose
or deoxyribose) a nitrogen contaning base (single
ringed pyrimidine or double ringed purine) and one or more phosphate
groups.
There are
three kind of nucleotides are the adenosine phosphates, the nucleotide
coenzymes and the nucleic acids ( deoxyribonucleic acid or DNA ribonucleic
acids or RNA).
Adenosine
Triphosphate (ATP):
ATP is
nucleotide that contain ribose (The sugar unit) adenine ( The base) and three
phosphate groups attached to the ribose.
It is
some time known as the energy currency of the body, which implies that the body
has to earn (Synthesize) it before it can send it. Many of the body`s huge
number of reactions release energy e.g. the breakdown of sugars in the presence
of Oxygen. The body captures energy released
by this reactions, using it to make ATP from adenosine diphosphate
(ADP). When the body needs chemical energy to fuel cellular activity, ATP
releases its stored energy and a
phosphate group through the splitting of a high energy phosphate bond, and
reverts to ADP. The body needs Chemical energy to:
1) Drive
synthetic reactions(Building Biological molecules).
2) Fuel
Movements.
3)
Transport substance across membrane.
 |
| Brief 3D Pictorial Description Post on Nucleotides |
Here are some Important Proteins structure in 3D (Through X-ray crystallography and NMR spectroscopy) From Protein Data Bank (PDB) which is made by European institute of bioinformatics.
1) Glycogen Phosphorylase:
 |
| Glycogen Phosphorylase: |
Although it may not seem so during the holiday season, we do not have to eat continually throughout the day. Our cells do require a constant supply of sugars and other nourishment, but fortunately our bodies contain a mechanism for storing sugar during meals and then metering it out for the rest of the day. The sugars are stored in glycogen, a large molecule that contains up to 10,000 glucose molecules connected in a dense ball of branching chains. Your muscles store enough glycogen to power your daily activities, and your liver stores enough to feed your nervous system and other tissues all through the day and on through the night.
Sweet Tooth
Sugar is released from glycogen by the enzyme glycogen phosphorylase. It clips glucose from the chains on the surface of a glycogen granule. The enzyme is a dimer of two identical subunits (colored green and blue in the structure here,In the upper illustration, two nucleotides, in red, are bound in the active site, which is found in a deep cleft. The yellow molecules are short chains of sugars similar to the ends of glycogen chains, which bind into another cleft that the enzyme uses to grip the glycogen granule. In its cleavage reaction, glycogen phosphorylase uses a phosphate molecule, connecting it to the sugar as it is released. A second enzyme, phosphoglucomutase, then shifts the position of the phosphate to a neighboring carbon atom in the sugar, making the sugar ready for breakdown by glycolysis.
Moderation
As you might imagine, this process is highly regulated. Traffic of sugar into and out of storage in glycogen is used to control the level of glucose in the blood, so glycogen phosphorylase must be activated when sugar is needed and quickly shut down when sugar is plentiful. It is controlled in several ways. First, the enzyme is activated by adding a phosphate molecule to a serine amino acid (serine 14) on the back side of the enzyme, shown in bright green and blue in the lower illustration. The phosphate causes a large shift in the shape of the enzyme (shown on the next page), shifting it into the active conformation. Two special enzymes control the addition and removal of this phosphate, based on levels of the sugar-monitoring hormones insulin and glucagon, and other hormones such as epinephrine (adrenaline).
Also, binding of other molecules can modify the activity of the molecule. For instance, AMP (adenine monophosphate) binds to a different site on the back side of the molecule (shown in red on the lower illustration), causing the same shift to the active conformation. This is useful, because AMP is a product of ATP breakdown and will be more plentiful when energy levels are low and more sugar is needed.
Glycogen Phosphorylase
 |
| Glycogen phosphorylase is activated by a change of shape |
Shape-shifting
Glycogen phosphorylase is activated by a change of shape. The structure on the left is in the inactive T state and the structure on the right is in the active R state. Compared with the pictures on the previous page, we are looking from the side and the active sites are on the left-hand side. (T stands for tense and R for relaxed, a notation developed when the first allosteric enzymes were being studied, although structures such as these have shown that the idea of tension does not really apply at the molecular level). The shift between the two shapes is controlled by phosphorylation of serine 14 or binding of AMP to the regulatory site. The R-state structure shown here has phosphates attached to the serines (colored pink) and a sulfate group in the site that binds to AMP (colored yellow).
Glycogen Phosphorylase
 |
| Glycogen is used in many organisms, from humans to yeast |
Exploring the Structure
Glycogen is used in many organisms, from humans to yeast. Much of the scientific work on the enzyme has been done with rabbit glycogen phosphorylase, which was shown on the previous two pages. You can look at the slightly different enzyme from yeast contains the two protein chains (colored blue and green here) and several small molecules. The molecule labeled PLP is the cofactor pyridoxal phosphate, a reactive molecule which binds tightly in the active site and is used to assist in the reaction. A phosphate is bound in each subunit next to the key threonine amino acid that is used for regulation, controlling an allosteric change similarly to serine 14 in the rabbit form. As you are looking at this enzyme, notice how the two protein chains wrap arms around one another. This allows the subunits to work together when responding to the small changes in shape that are used for control.
2) Ribosome
 |
| Ribosome |
The Protein Factory
Protein synthesis is the major task performed by living cells. For instance, roughly one third of the molecules in a typical bacterial cell are dedicated to this central task. Protein synthesis is a complex process involving many molecular machines. You can look at many of these molecules in the PDB, including DNA, DNA polymerases, and RNA polymerases; a host of repressors, DNA repair enzymes, topoisomerases, and histones; tRNA and acyl-tRNA synthetases; and molecular chaperones. This month, for the first time, you can also look at the factory of protein synthesis in atomic detail.
An Elusive Structure
The ribosome has been under the scrutiny of scientists for decades. Electron microscopy has yielded an increasingly detailed view over the years, defining the overall shape of individual ribosomes and differences in this shape for ribosomes from different species, More recently, detailed electron micrograph reconstructions have studied the interaction or ribosomes with messenger RNA, transfer RNA and the protein elongation factors. This legacy of morphological work lays the groundwork on which the atomic structures may be understood.
Ribosomes are composed of two subunits: a large subunit, shown on the right, and a small subunit, shown on the left. Of course, the term "small" is used in a relative sense here: both the large and the small subunits are huge compared to a typical protein. Both subunits are composed of long strands of RNA, shown here in orange and yellow, dotted with protein chains, shown in blue. When synthesizing a new protein, the two subunits lock together with a messenger RNA trapped in the space between. The ribosome then walks down the messenger RNA three nucleotides at a time, building a new protein piece-by-piece.
Ribosome : The Large Subunit
 |
| Ribosome : The Large Subunit |
The structure of the large subunit is available in . The large subunit contains the active site of the ribosome: the site that creates the new peptide bonds when proteins are synthesized. In this view, the messenger RNA would run horizontally in the groove across the middle. This structure, along with several other structures with inhibitors bound, provide strong evidence that the ribosome is a ribozyme. Enzymes typically use amino acids to catalyze chemical reactions, but the ribosome appears to use an adenine RNA nucleotide to perform its synthetic task. This adenine is colored green in the figure (we'll look at this more closely later).
The large subunit is composed of two RNA strands: a long one colored orange and a shorter one colored yellow. Dozens of proteins bind on the surface of the ribosome. Many have long, snaky tails that extend into the body of the ribosome, gluing the RNA strands into their proper shape. Several of the proteins were not seen in this crystallographic structure, perhaps because they are too flexible. Approximate shapes for these proteins, which form two prominent stalks commonly used as landmarks in electron micrographs, are indicated here in light blue.
Ribosome :The Small Subunit
 |
| Ribosome :The Small Subunit |
The structure of the small subunit is available in the The small subunit is in charge of information flow during protein synthesis. It initially finds a messenger RNA strand and, after combining with a large subunit, ensures that each codon in the message is paired with the anticodon in the proper transfer RNA. The messenger RNA is thought to enter through a small hole (seen here on the left side of the molecule) and then extend up into the "decoding center" in the cleft between the "head" at top and the "body" at the bottom. The messenger RNA does not have to thread through this hole like a needle, however, because the hole is actually formed by a loop of the ribosomal RNA, which can open like a latch to admit the messenger.
 |
| Exploring the Structures |
Exploring the Structures
Before jumping into these structures, be prepared. Both the large subunit and the small subunit are enormous complexes with many atoms: the structure of the large subunit in contains over 64,000 atoms, even though the authors chose to release only alpha carbon positions for the proteins, and the small subunit structure also with partial structures for the proteins, contains almost 35,000 atoms. Many interactive display programs become very sluggish when working on structures this large.
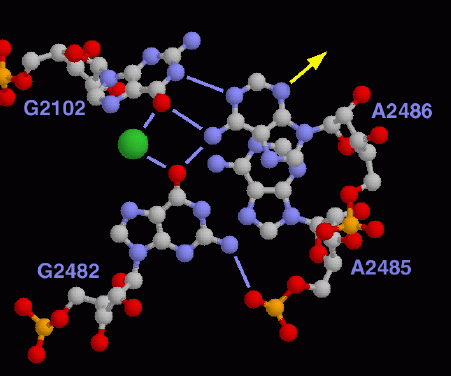
The picture shown here shows the proposed active site in the large ribosomal subunit. Adenine 2486 is thought to perform the synthesis reaction, at the location indicated by the yellow arrow. The two guanines shown on the left and the potassium ion shown in green serve to activate this adenine through a series of hydrogen bonds, shown in light blue. This picture was created with RasMol. You can create a similar picture by turning off display of the whole structure and then selectively displaying A2486, A2485, G2102, and G2482.
3) Restriction Enzymes
 |
| Restriction Enzymes |
Bacteria Fight Back
Bacteria are under constant attack by bacteriophages, like the bacteriophage phiX174 described in an earlier Molecule of the Month. To protect themselves, many types of bacteria have developed a method to chop up any foreign DNA, such as that of an attacking phage. These bacteria build an endonuclease--an enzyme that cuts DNA--which is allowed to circulate in the bacterial cytoplasm, waiting for phage DNA. The endonucleases are termed "restriction enzymes" because they restrict the infection of bacteriophages.
Molecular Scissors
Each type of restriction enzyme seeks out a single DNA sequence and precisely cuts it in one place. For instance, the enzyme shown here, EcoRI, cuts the sequence GAATTC, cutting between the G and the A. Of course, roving endonucleases can be dangerous, so bacteria protect their own DNA by modifying it with methyl groups. These groups are added to adenine or cytosine bases (depending on the particular type of bacteria) in the major groove. The methyl groups block the binding of restriction enzymes, but they do not block the normal reading and replication of the genomic information stored in the DNA. DNA from an attacking bacteriophage will not have these protective methyl groups and will be destroyed. Each particular type of bacteria has a restriction enzyme (or several different ones) that cuts a specific DNA sequence, paired with a methyl-transferase enzyme that protects this same sequence in the bacterial genome.
Restriction Enzymes : Sticky Ends
 |
The booming field of biotechnology was made possible by the discovery of restriction enzymes in the early 1950's |
The booming field of biotechnology was made possible by the discovery of restriction enzymes in the early 1950's. With them, DNA may be cut in precise locations. A second enzyme--DNA ligase--may then be used to reassemble the pieces in any desired order. Together, these two enzymes allow researchers to assemble customized genomes. For instance, researchers can create designer bacteria that make insulin or growth hormone or add genes for disease resistance to agricultural plants.
An interesting property of restriction enzymes simplifies this molecular cutting and pasting. Restriction enzymes typically recognize a symmetrical sequence of DNA, Notice that the top strand is the same as the bottom strand, read backwards. When the enzyme cuts the strand between G and A, it leaves overhanging chains:These are termed "sticky ends" because the base pairs formed between the two overhanging portions will glue the two pieces together, even though the backbone is cut. Sticky ends are an essential part of genetic engineering, allowing researchers to cut out little pieces of DNA and place them in specific places, where the sticky ends match.
 |
| Another example from Escherichia coli--EcoRV--is shown here |
Exploring the Structure
The PDB contains structures for many restriction enzymes. Another example from Escherichia coli--EcoRV--is shown here. The structure at the top, shows the enzyme bound to a short piece of DNA. The arrow shows the phosphate group that will be cut. The lower illustration, taken from shows the structure after the DNA has been cut. A water molecule has been inserted, so there are now two oxygen atoms, close to one another but not bonded together, where there was a single bonded oxygen atom in the intact DNA.
























 Online Movies
Online Movies
This comment has been removed by the author.
ReplyDeleteSure , and thank you for sharing.
ReplyDeleteCustom peptide - The information that you provided was helpful for readers. I will have to share your article with others.
ReplyDelete