 |
| Protein Post |
Proteins
and Amino Acids:
Amino
acids always contain Carbon, Hydrogen, Oxygen and Nitrogen, and many in
addition carry Sulphur. In human Bio-Chemistry, 20 Amino acids are used as the
principal building blocks of protein, although there are other; for instance,
there are some amino-acids used only in certain proteins and some are seen only
in microbial products. Of The amino acids used in human protein synthesis,
there is a basic common structure, including an amino
group (NH2) a carboxyl
group (COOH) and a hydrogen atom. What makes one amino acid different from
next is a variable side chain. As in formation of glycosidic linkage, when two
amino acids join up the reaction expels a molecule of water and the resulting
bond is called a peptide bond.
Proteins
are made from amino acids joined together, and are the main family of molecules
from which the human body is built. Protein Molecules vary enormously in size,
shape, chemical constituents and function. Many important groups of
Biologically active substance are proteins:
1)
Carrier molecules e.g. Haemoglobin.
2)
Enzymes.
3) Many
hormones e.g. Insulin.
4)
Antibodies.
5) Motor
Protein
6)
Receptor Protein e.g Rhodopsin
7)
Structural Proteins
8)
Storage Proteins
etc.
Proteins
can also be used as an alternative energy source, usually in dietary
inadequancey, although the process in much less efficient than when
Carbohydrates or fats are broken down.
Primary
Structure of Proteins:
A
covalent bond forms between the amino group of one amino acid and the carboxyl
group to another. This covalent linkage, called a peptide bond, results in a
molecule called dipeptide. Three or more amino acids linked together form
polypeptide chain. Which kind of amino
acid follows another in the chain is always the same for all proteins of a
given type. For example, the two chains making up the protein Insulin always
have the sequences same. The specific sequence of amino acids in a polypeptide
chain constitute the primary structure of protein.
Secondery
Structure of Proteins:
The term
refers to the helical or extended pattern brought about by Hydrogen bonds at
regular intervals along a polypeptide chain. Some amino acids tend to favour
helical pattern, other tend to favour sheetlike patterns.
Tertiary
Structure of Proteins:
Protein
structure is also affected by interactions among R groups. Most helically
coiled chains become further folded into some characteristic shape when one R
group interacts with another R group some distance away, with the backbone
itself, or with other substances present in the cell. The term tertiary
structure refers to the folding that arises through interaction among R groups
of a polypeptide chain.
Quaternary
Structure of protein:
The 4th
level of protein in architecture, results from interactions between two or more
polypeptide chains in some protein. The resulting protein can be
globular,fiberlike,or some combination of the two shapes e.g. Haemoglobin.
 |
| Brief Pictorial Summery of Proteins |
Structure
determination
Discovering
the tertiary structure of a protein, or the quaternary structure of its
complexes, can provide important clues about how the protein performs its
function. Common experimental methods of structure determination include X-ray
crystallography and NMR spectroscopy, both of which can produce information at
atomic resolution. However, NMR experiments are able to provide information
from which a subset of distances between pairs of atoms can be estimated, and
the final possible conformations for a protein are determined by solving a
distance geometry problem. Dual polarisation interferometry is a quantitative
analytical method for measuring the overall protein conformation and
conformational changes due to interactions or other stimulus. Circular
dichroism is another laboratory technique for determining internal beta sheet/
helical composition of proteins. Cryoelectron microscopy is used to produce
lower-resolution structural information about very large protein complexes,
including assembled viruses; a variant known as electron crystallography can
also produce high-resolution information in some cases, especially for
two-dimensional crystals of membrane proteins. Solved structures are usually
deposited in the Protein Data Bank (PDB), a freely available resource from
which structural data about thousands of proteins can be obtained in the form
of Cartesian coordinates for each atom in the protein.
Many more
gene sequences are known than protein structures. Further, the set of solved
structures is biased toward proteins that can be easily subjected to the
conditions required in X-ray crystallography, one of the major structure
determination methods. In particular, globular proteins are comparatively easy
to crystallize in preparation for X-ray crystallography. Membrane proteins, by
contrast, are difficult to crystallize and are underrepresented in the PDB.
Structural genomics initiatives have attempted to remedy these deficiencies by
systematically solving representative structures of major fold classes. Protein
structure prediction methods attempt to provide a means of generating a plausible
structure for proteins whose structures have not been experimentally
determined.
Different Types of Proteins :
 |
| 1) Enzyme Protein |
 |
| 2) Gene Regulatory Protein |
 |
| 3) Motor Proteins |
 |
| 4) Receptor Protein |
 |
| 5) Signaling Proteins |
 |
| 6) Proteins For Special Purposes |
 |
| 7) Storage Proteins |
 |
| 8) Structural Protein |
 |
| 9) Transport Protein |
Nucleotides:
These are
the largest molecules in the body and are built from components called
nucleotides, which consists of three subunits are : A sugar ( ribose
or deoxyribose) a nitrogen contaning base (single
ringed pyrimidine or double ringed purine) and one or more phosphate
groups.
There are
three kind of nucleotides are the adenosine phosphates, the nucleotide
coenzymes and the nucleic acids ( deoxyribonucleic acid or DNA ribonucleic
acids or RNA).
Adenosine
Triphosphate (ATP):
ATP is
nucleotide that contain ribose (The sugar unit) adenine ( The base) and three
phosphate groups attached to the ribose.
It is
some time known as the energy currency of the body, which implies that the body
has to earn (Synthesize) it before it can send it. Many of the body`s huge
number of reactions release energy e.g. the breakdown of sugars in the presence
of Oxygen. The body captures energy released
by this reactions, using it to make ATP from adenosine diphosphate
(ADP). When the body needs chemical energy to fuel cellular activity, ATP
releases its stored energy and a
phosphate group through the splitting of a high energy phosphate bond, and
reverts to ADP. The body needs Chemical energy to:
1) Drive
synthetic reactions(Building Biological molecules).
2) Fuel
Movements.
3)
Transport substance across membrane.
 |
| Brief 3D Pictorial Description Post on Nucleotides |
Here are some Important Proteins structure in 3D (Through X-ray crystallography and NMR spectroscopy) From Protein Data Bank (PDB) which is made by European institute of bioinformatics.
Insulin:
 |
| Insulin : A Molecular Messenger |
A Molecular Messenger:
Our cells communicate using a molecular postal system: the blood is the postal service and hormones are the letters. Insulin is one of the most important hormones, carrying messages that describe the amount of sugar that is available from moment to moment in the blood. Insulin is made in the pancreas and added to the blood after meals when sugar levels are high. This signal then spreads throughout the body, to the liver, muscles and fat cells. Insulin tells these organs to take glucose out of the blood and store it, in the form of glycogen or fat.
Folding Tiny Proteins:
Insulin is a tiny protein. It moves quickly through the blood and is easily captured by receptors on cell surfaces, delivering its message. Small proteins pose a challenge to cells: it is difficult to make a small protein that will fold into a stable structure. Our cells solve this problem by synthesizing a longer protein chain, which folds into the proper structure. Then, the extra piece is clipped away, leaving two small chains in the mature form. These two chains are shown in the lower diagram in blue and green, for insulin from pigs .The structure is further stabilized by three disulfide bridges, one of which is seen in yellow in each illustration.
Diabetes Mellitus:
When insulin function is impaired, either by damage to the pancreas or by the rigors of aging, glucose levels in the blood rise dangerously, leading to diabetes mellitus. For people totally deficient in insulin, such as children that develop diabetes early in life, this can be acutely dangerous. High glucose levels lead to dehydration, as the body attempts to flush out the excess sugar in urine, and life-threatening changes in blood pH, as the body turns to other acidic molecules for delivery of energy. Diabetes mellitus has severe long- term effects as well. It is one of the major chronic diseases in the industrialized world. Lowered levels of insulin that may occur as we age allow elevated levels of sugar in the blood over extended periods of time. Sugar molecules attach to proteins throughout the body, compromising their function, and sugars derived from glucose build up, distorting and clogging cells.
 |
| A Molecular Messenger |
Insulin Therapy:
Diabetes mellitus may be treated by manually replacing the insulin that is missing in the blood. Of course, we need a plentiful source of insulin for use in these treatments. Fortunately, insulin from pigs differs from human insulin by only one amino acid: a threonine at the end of the chain in human insulin is replaced by alanine in pig insulin. Insulin from cows is also very similar, differing in only three positions. Because of their similarity, these forms of insulin are also recognized by our own cells and may be used in therapy. Today, human insulin is also created by biotechnology, using engineered bacteria to produce a protein exactly identical to our own protein.
 |
| Exploring the Structure |
Exploring the Structure:
Insulin is a perfect molecule for exploring protein structure. It is small enough that you can display all of the atoms and still have a picture that is not too confusing. Human insulin is pictured here. The file contains four chains, labeled A, B, C, and D. When looking at this structure yourself, you will want to display only the A and B chains, which together compose one monomer of insulin. In the structure, you can see many of the key features that stabilize protein structure. Notice the cluster of carbon-rich amino acids, like leucine and isoleucine, that cluster in the middle of insulin, forming a hydrophobic core. Notice that the surface is covered with the charged amino acids lysine, arginine, and glutamate. These amino acids interact favorably with the surrounding water. Also notice the three disulfide bridges between cysteine amino acids, which stabilize this tiny protein.
2) Antibodies
 |
| Antibody |
Antibodies are our molecular watchdogs, waiting and watching for viruses, bacteria and other unwelcome visitors. Antibodies circulate in the blood, scrutinizing every object that they touch. When they find an unfamiliar, foreign object, they bind tightly to its surface. In the case of viruses, like rhinovirus or poliovirus , a coating of bound antibodies may be enough to block infection. Antibodies alone, however, are no match for bacteria. When antibodies bind to a bacterial surface, they act as markers alerting the other powerful defensive mechanisms available in the immune system.
Getting a Grip:
Antibodies, and many of the other molecules used in the immune system, have a distinctive shape. Typically, they are composed of several flexible arms with binding sites at the end of each one. This make perfect sense: since antibodies do not know in advance what attackers they might be fighting, they keep their options open. The flexible arms allow the binding sites to work together, grabbing with both arms onto targets with different overall shapes. has two binding sites, at the tips of the two arms extending right and left at the top. Notice the thin, flexible chains that connect these arms to the central domain at the bottom. Some antibodies have longer flexible linkers connecting the arms together, allowing them even more latitude when finding purchase on a surface. Other antibodies have four or ten binding sites, so each contact can be weaker and still allow the whole antibody to bind firmly.
 |
| Your blood contains upwards of 100,000,000 different types of antibodies |
Power in Numbers:
Your blood contains upwards of 100,000,000 different types of antibodies. Each type binds to a different target molecule. Remarkably, all of these antibodies are created before they ever see a virus or bacterium. You don't make a special antibody when a virus or bacterium infects your body. Instead, all of your antibodies are pre-fabricated, lying in wait until a virus or bacterium attacks. There are so many different kinds of antibodies that one or two are bound to be the right ones to fight the infection.
This amazingly huge collection of antibodies is created by recombination of genes in lymphocytes, the blood cells that make antibodies. Each lymphocyte creates a different type of antibody, based on how it has recombined its antibody genes. When an antibody encounters a virus or bacterium, the appropriate lymphocytes will multiply, flooding the blood with the particular antibodies needed to battle the invader. These lymphocytes may also make small adjustments on the antibodies they produce, tailoring their antibodies to bind more tightly and more specifically.
Antibody Structure:
Antibodies are composed of four chains, two long heavy chains (colored red and orange) and two shorter light chains (yellow). The specific binding site is found at the tips of the two arms, in a pocket formed between the light and heavy chain. The binding site is composed of several loops in the protein chain that have very different lengths and amino acid composition. Differences in these "hypervariable loops" form the many types of pockets in different antibodies, each of which bind specifically to a different target. The rest of the antibody--the rest of the arms and the large constant domain that ties the two arms together--is relatively uniform in structure, providing a convenient handle when antibodies interact with the rest of the immune system.
 |
| Antibody Structure |
Attack From Many Angles:
When a foreign molecule is found in the blood, many different antibodies may bind to it, attacking at different angles. Three different antibodies that bind to the protein lysozyme (in green at the center) are shown here. The crystal structures each include only one arm of the antibody (termed "Fab" for "antigen-binding fragment"), which has been separated from the antibody for ease in study. The rest of the antibody is indicated extending from the edges of the illustration. Notice that the antibodies pick entirely different binding sites on the small lysozyme molecule.
3) DNA Ligase
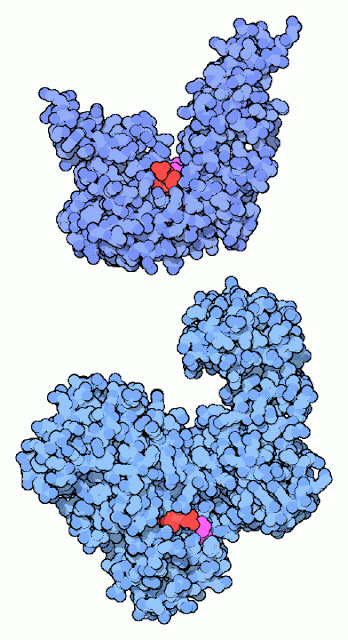 |
| DNA-Ligase |
Human cells (with a few unusual exceptions) each contain their own set of 46 long strands of DNA. All of our genetic information is encoded in these strands, with thousands of genes strung along their length. The ordering of genes, and the proximity of one next to the other, can be important for the proper usage of the information, so it is important that our cells protect their DNA from breakage. If one strand in the DNA breaks, it is not a disaster, but it can lead to problems when the DNA double helix is unwound during the processes of transcription and replication. Breakage of both strands, on the other hand, is far more serious. To protect us from these dangers, our cells use DNA ligases to glue together DNA strands that have been broken.
Breaking DNA:
Environmental hazards can accidentally damage DNA. Ionizing radiation, such as gamma rays, attack the backbone of DNA and cause breaks. Our cells are also bathed with oxygen, a dangerous gas that forms reactive free radicals that attack DNA. Surprisingly, our cells also break their own DNA on purpose. During meiosis, the process where the genome is split into two halves for creation of egg or sperm cells, the DNA is often recombined. Portions of one DNA strand are cut out and traded with similar portions on a sister chromosome. Cells of the immune system also shuffle around their DNA strands, cutting out pieces and replacing them in a different location to build up a diverse collection of antibody genes. Gaps--breaks in one strand of the DNA--are also created when the DNA is replicated. Polymerases only work in one direction when they copy a DNA strand, so one of the two strands is copied in a series of small pieces that need to be linked up into a continuous strand.
Repairing Broken DNA:
DNA ligase reconnects DNA strands when they are broken. It uses a cofactor molecule (shown in red) for power and a special lysine amino acid (shown in magenta) to perform the reaction. Our DNA ligases and the DNA ligase from the bacteriophage T7 use ATP as the cofactor. Many bacteria, on the other hand, use NAD in the reaction, as with the DNA ligase shown at the bottom (from PDB entry In both cases, a lysine in the DNA ligase forms a bond to the phosphate in the cofactor, holding onto the AMP portion and discarding the rest. Later in the reaction, this AMP is transferred to the broken DNA strand, and then is released when the strand is rejoined.
Gene Jockeys:
DNA ligases are remarkably useful enzymes, inside cells and out. Along with restriction enzymes they have made the entire field of recombinant DNA technology possible. With these two types of enzymes, researchers can cut and paste DNA strands at will, designing new genes and new genomes. Restriction enzymes are like scissors, allowing us to cut DNA in specific places. DNA ligase then allows us to reconnect them into functional DNA strands. With this technology, we are now able to do for ourselves what cells have been doing for billions of years!
Non-Homologous End Joining:
 |
| One of the important reactions performed by DNA ligase is non-homologous end joining |
One of the important reactions performed by DNA ligase is non-homologous end joining. This occurs when both strands of the DNA break, and the cell has to glue it back together. It is an emergency repair system, so it may introduce an error or two in the genetic information as the two ends are prepared for rejoining, but that is certainly better than leaving the DNA in fragments. A few of the molecules involved in the process are shown here. The Ku protein, and a DNA-dependent protein kinase (not shown) are thought to bring the two ends together and hold them in place. Notice how the two subunits of Ku have arms that wrap around the DNA strand. DNA ligase, assisted by the Xrcc4 protein seal the strands back together. The DNA ligase is shown as a schematic, since the atomic structure of the human form has not yet been solved.
Exploring the Structure
 |
| Exploring the Structure of DNA Ligase |
you can take a close look at the activated AMP molecule used in the ligation reaction. This ligase, from Chorella virus, is a smaller version of the one found in our cells. Lysine 27 forms a covalent bond with the phosphate of the AMP, making it ready for the ligation reaction. The DNA will bind in the large groove at the top.
























 Online Movies
Online Movies
No comments:
Post a Comment