Proteins
and Amino Acids:
Amino
acids always contain Carbon, Hydrogen, Oxygen and Nitrogen, and many in
addition carry Sulphur. In human Bio-Chemistry, 20 Amino acids are used as the
principal building blocks of protein, although there are other; for instance,
there are some amino-acids used only in certain proteins and some are seen only
in microbial products. Of The amino acids used in human protein synthesis,
there is a basic common structure, including an amino
group (NH2) a carboxyl
group (COOH) and a hydrogen atom. What makes one amino acid different from
next is a variable side chain. As in formation of glycosidic linkage, when two
amino acids join up the reaction expels a molecule of water and the resulting
bond is called a peptide bond.
Proteins
are made from amino acids joined together, and are the main family of molecules
from which the human body is built. Protein Molecules vary enormously in size,
shape, chemical constituents and function. Many important groups of
Biologically active substance are proteins:
1)
Carrier molecules e.g. Haemoglobin.
2)
Enzymes.
3) Many
hormones e.g. Insulin.
4)
Antibodies.
5) Motor
Protein
6)
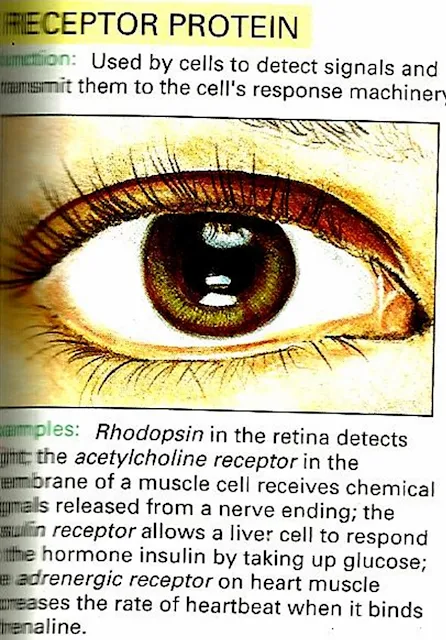
Receptor Protein e.g Rhodopsin
7)
Structural Proteins
8)
Storage Proteins
etc.
Proteins
can also be used as an alternative energy source, usually in dietary
inadequancey, although the process in much less efficient than when
Carbohydrates or fats are broken down.
Primary
Structure of Proteins:
A
covalent bond forms between the amino group of one amino acid and the carboxyl
group to another. This covalent linkage, called a peptide bond, results in a
molecule called dipeptide. Three or more amino acids linked together form
polypeptide chain. Which kind of amino
acid follows another in the chain is always the same for all proteins of a
given type. For example, the two chains making up the protein Insulin always
have the sequences same. The specific sequence of amino acids in a polypeptide
chain constitute the primary structure of protein.
Secondery
Structure of Proteins:
The term
refers to the helical or extended pattern brought about by Hydrogen bonds at
regular intervals along a polypeptide chain. Some amino acids tend to favour
helical pattern, other tend to favour sheetlike patterns.
Tertiary
Structure of Proteins:
Protein
structure is also affected by interactions among R groups. Most helically
coiled chains become further folded into some characteristic shape when one R
group interacts with another R group some distance away, with the backbone
itself, or with other substances present in the cell. The term tertiary
structure refers to the folding that arises through interaction among R groups
of a polypeptide chain.
Quaternary
Structure of protein:
The 4th
level of protein in architecture, results from interactions between two or more
polypeptide chains in some protein. The resulting protein can be
globular,fiberlike,or some combination of the two shapes e.g. Haemoglobin.
 |
| Brief Pictorial Summery of Proteins |
Structure
determination
Discovering
the tertiary structure of a protein, or the quaternary structure of its
complexes, can provide important clues about how the protein performs its
function. Common experimental methods of structure determination include X-ray
crystallography and NMR spectroscopy, both of which can produce information at
atomic resolution. However, NMR experiments are able to provide information
from which a subset of distances between pairs of atoms can be estimated, and
the final possible conformations for a protein are determined by solving a
distance geometry problem. Dual polarisation interferometry is a quantitative
analytical method for measuring the overall protein conformation and
conformational changes due to interactions or other stimulus. Circular
dichroism is another laboratory technique for determining internal beta sheet/
helical composition of proteins. Cryoelectron microscopy is used to produce
lower-resolution structural information about very large protein complexes,
including assembled viruses; a variant known as electron crystallography can
also produce high-resolution information in some cases, especially for
two-dimensional crystals of membrane proteins. Solved structures are usually
deposited in the Protein Data Bank (PDB), a freely available resource from
which structural data about thousands of proteins can be obtained in the form
of Cartesian coordinates for each atom in the protein.
Many more
gene sequences are known than protein structures. Further, the set of solved
structures is biased toward proteins that can be easily subjected to the
conditions required in X-ray crystallography, one of the major structure
determination methods. In particular, globular proteins are comparatively easy
to crystallize in preparation for X-ray crystallography. Membrane proteins, by
contrast, are difficult to crystallize and are underrepresented in the PDB.
Structural genomics initiatives have attempted to remedy these deficiencies by
systematically solving representative structures of major fold classes. Protein
structure prediction methods attempt to provide a means of generating a plausible
structure for proteins whose structures have not been experimentally
determined.
Different Types of Proteins :
 |
| 1) Enzyme Protein |
 |
| 2) Gene Regulatory Protein |
 |
| 3) Motor Proteins |
 |
| 4) Receptor Protein |
 |
| 5) Signaling Proteins |
 |
| 6) Proteins For Special Purposes |
 |
| 7) Storage Proteins |
 |
| 8) Structural Protein |
 |
| 9) Transport Protein |
Nucleotides:
These are
the largest molecules in the body and are built from components called
nucleotides, which consists of three subunits are : A sugar ( ribose
or deoxyribose) a nitrogen contaning base (single
ringed pyrimidine or double ringed purine) and one or more phosphate
groups.
There are
three kind of nucleotides are the adenosine phosphates, the nucleotide
coenzymes and the nucleic acids ( deoxyribonucleic acid or DNA ribonucleic
acids or RNA).
Adenosine
Triphosphate (ATP):
ATP is
nucleotide that contain ribose (The sugar unit) adenine ( The base) and three
phosphate groups attached to the ribose.
It is
some time known as the energy currency of the body, which implies that the body
has to earn (Synthesize) it before it can send it. Many of the body`s huge
number of reactions release energy e.g. the breakdown of sugars in the presence
of Oxygen. The body captures energy released
by this reactions, using it to make ATP from adenosine diphosphate
(ADP). When the body needs chemical energy to fuel cellular activity, ATP
releases its stored energy and a
phosphate group through the splitting of a high energy phosphate bond, and
reverts to ADP. The body needs Chemical energy to:
1) Drive
synthetic reactions(Building Biological molecules).
2) Fuel
Movements.
3)
Transport substance across membrane.
 |
| Brief Pictorial Summery of Nucleotides |
Here are
some Important Proteins structure in 3D
(Through X-ray crystallography and NMR spectroscopy) From Protein Data
Bank (PDB) which is made by European institute of bioinformatics.
Your Most Plentiful Protein
About
one quarter of all of the protein in your body is collagen. Collagen is a major
structural protein, forming molecular cables that strengthen the tendons and
vast, resilient sheets that support the skin and internal organs. Bones and
teeth are made by adding mineral crystals to collagen. Collagen provides
structure to our bodies, protecting and supporting the softer tissues and
connecting them with the skeleton. But, in spite of its critical function in
the body, collagen is a relatively simple protein.
The Collagen Triple Helix
Collagen
is composed of three chains, wound together in a tight triple helix. The
illustration included here shows only a small segment of the entire
molecule--each chain is over 1400 amino acids long and only about 20 are shown
here. A repeated sequence of three amino acids forms this sturdy structure.
Every third amino acid is glycine, a small amino acid that fits perfectly
inside the helix. Many of the remaining positions in the chain are filled by
two unexpected amino acids: proline and a modified version of proline,
hydroxyproline. We wouldn't expect proline to be this common, because it forms
a kink in the polypeptide chain that is difficult to accommodate in typical
globular proteins. But, as you can see on the next page, it seems to be just
the right shape for this structural protein.
Vitamin C
Hydroxyproline,
which is critical for collagen stability, is created by modifying normal
proline amino acids after the collagen chain is built. The reaction requires
vitamin C to assist in the addition of oxygen. Unfortunately, we cannot make
vitamin C within our bodies, and if we don't get enough in our diet, the
results can be disastrous. Vitamin C deficiency slows the production of
hydroxyproline and stops the construction of new collagen, ultimately causing
scurvy. The symptoms of scurvy--loss of teeth and easy bruising-- are caused by
the lack of collagen to repair the wear-and-tear caused by everyday activities.
Collagen on the Grocery Shelf
Collagen
from livestock animals is a familiar ingredient for cooking. Like most
proteins, when collagen is heated, it loses all of its structure. The triple
helix unwinds and the chains separate. Then, when this denatured mass of tangled
chains cools down, it soaks up all of the surrounding water like a sponge,
forming gelatin.
Collagen
Exploring
the Structure :
 |
| 3D Structure of Collagen |
A
special amino acid sequence makes the tight collagen triple helix particularly
stable. Every third amino acid is a glycine, and many of the remaining amino
acids are proline or hydroxyproline. A classic triple helix is shown here,
Notice how the glycine forms a tiny elbow packed inside the helix, and notice
how the proline and hydroxyproline smoothly bend the chain back around the
helix. In this structure, the researchers placed a larger alanine amino acid in
the position normally occupied by glycine, showing that it crowds the
neighboring chains.
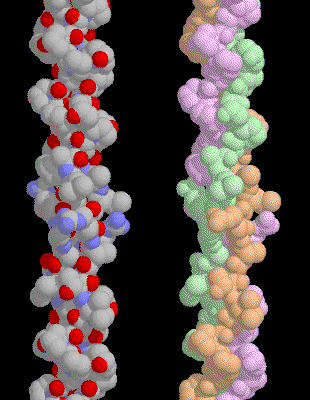 |
| This collagen helix contains a segment of human collagen, Notice that the top half is very uniform |
This
collagen helix contains a segment of human collagen, Notice that the top half
is very uniform, where the sequence is the ideal mixture of glycine and
prolines. At the bottom, the helix is less regular, because many different
amino acids are placed between the equally-spaced glycines.
2) ATP Synthase:
 |
| 3D Picture of ATP Synthase |
ATP synthase is one of the wonders of the molecular world. ATP synthase is an enzyme, a molecular motor, an ion pump, and another molecular motor all wrapped together in one amazing nanoscale machine. It plays an indispensable role in our cells, building most of the ATP that powers our cellular processes. The mechanism by which it performs this task is a real surprise.
Rotary Motors:
ATP synthesis is composed of two rotary motors, each powered by a different fuel. The motor at the top, termed F0, an electric motor. It is embedded in a membrane (shown schematically as a gray stripe here), and is powered by the flow of hydrogen ions across the membrane. As the protons flow through the motor, they turn a circular rotor (shown in blue). This rotor is connected to the second motor, termed F1. The F1 motor is a chemical motor, powered by ATP. The two motors are connected together by a stator, shown on the right, so that when F0 turns, F1 turns too.
Motor to Generator:
So why have two motors connected together? The trick is that one motor can force the other motor to turn, and in this way, change the motor into a generator. This is what happens in our cells: the F0 motor uses the power from a proton gradient to force the F1 motor to generate ATP. In our cells, food is broken down and used to pump hydrogen ions across the mitochondrial membrane. The F0 portion of ATP synthase allows these ions to flow back, turning the rotor in the process. As the rotor turns, it turns the axle and the F1 motor becomes a generator, creating ATP as it turns.
Parts List:
Large, complex molecular machines like ATP synthase pose difficult problems for structural scientists, so the structures of these machines are often determined in parts. The picture shown here is a composite of four different structures, combining structures determined by X-ray crystallography and NMR spectroscopy.
Exploring the F1 Structure:
 |
| Exploring the F1 Part Structure of ATP Synthase |
When operating as a generator, it uses the power of rotational motion to build ATP, or when operating as a motor, it breaks down ATP to spin the axle the opposite direction. The synthesis of ATP requires several steps, including the binding of ADP and phosphate, the formation of the new phosphate-phosphate bond, and release of ATP. As the axle turns, it forces the motor into three different conformations that assist these difficult steps. Two states are shown here. The one on the left shows a conformation that assists the binding of ADP, and the one on the right shows a conformation that has been forced open to release ATP. Notice how the oddly-shaped axle forces the change in conformation.
Exploring the F0 Structure:
 |
| Exploring the F0 Part Structure of ATP Synthase |
In this picture, we are looking down the axis of rotation, as if we where looking down at the top of the picture on the first page. The rotor is composed of 12 identical protein chains, colored blue here, and the ion pump is a single chain, colored red. The pump has an arginine amino acid that hands off a hydrogen ion to aspartates on the rotor. Aspartate amino acids typically have a negative charge, but since the rotor is surrounded by membrane lipids, this would be very unfavorable. So, the rotor only turns when the aspartates have a hydrogen attached, neutralizing their charge. Hydrogen ions take a convoluted path through the F0 motor, turning the rotor in the process. They are gathered by a chain of amino acids in the pump, and transferred to the arginine. The arginine passes the hydrogen to the rotor, which turns all the way around. Then the hydrogen is offloaded by other amino acids on the pump, and finally passed to the opposite side of the membrane. The exact path of the hydrogen ions through the pump is still a matter of intense study.
3) Circadian Clock Proteins :
 |
| 3D structure of Circadian Clock Proteins |
Our cells contain tiny molecular clocks that measure out a 24-hour circadian rhythm. This clock decides when we get hungry and when we get sleepy. This clock can sense when the days are getting longer and shorter, and then trigger seasonal changes. Our major clock is housed in a small region of the brain, called the suprachiasmic nuclei. It acts as our central pacemaker, checking the cycles of light and dark outside, and then sending signals to synchronize clocks throughout the rest of the body.
Counting the Hours :
Molecular processes occur so fast that is it difficult to imagine a 24-hour clock that works at the molecular level. But surprisingly, different organisms have evolved many different ways of doing this. Animal cells use a complex collection of proteins (with fanciful names like Clock, Cryptochrome, and Period) that are rhythmically synthesized and degraded each day. The 24-hour oscillation of the levels of these proteins is controlled by a series of interconnected feedback loops, where the levels of the proteins precisely regulate their own production. A much simpler system has been discovered in cyanobacteria. It is composed of three proteins, KaiA, KaiB and KaiC, that together form a circadian clock. At the beginning of the cycle, KaiA . Then, as KaiC fills itself up with phosphates, it binds to KaiB . As the number of phosphates drops, KaiB falls off and KaiA can start the cycle again.
Synchronize Your Watches :
These clocks have a period of about 24 hours, but as you can imagine, they are not exact. So cells have a way of synchronizing their clocks with the outside world. The clock in our brain is synchronized by exposure to light. Light is sensed by the retina, and signals are sent into the brain to modify the timing of the circadian oscillations. If you have traveled across several time zones, you have experienced this synchronization. For the first day or two, you experience jet lag because your clock is synchronized with the old schedule. But gradually, the bright light of day (blue light seems to work best) shifts your clock to bring you into alignment with the local time.
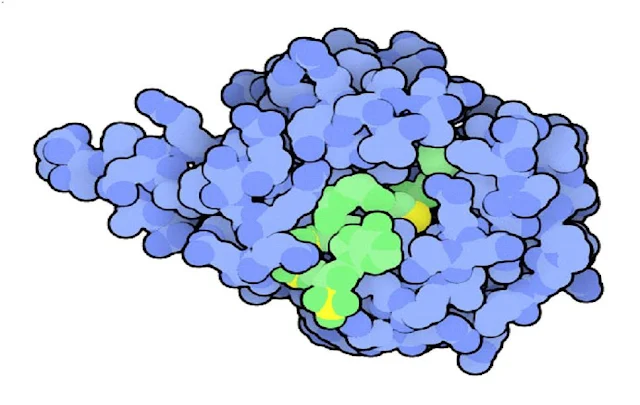 |
| Sleepy Molecules |
Sleepy Molecules :
The small hormone melatonin is produced selectively at night, and circulates through the blood to coordinate our nightly activities, such as sleep. Treatment with melatonin may be used to change this cycle artificially, for instance helping to shift cycles that are out of phase during jet lag. The daily rise and fall of melatonin is caused by changes in the levels of the enzyme serotonin N-acetyltransferase, shown here This enzyme adds a few atoms to the neurotransmitter serotonin, then a second enzyme converts it into melatonin. In this structure, the enzyme is caught in the process of performing the reaction--the large green molecule bound in the active site is similar to the intermediate formed as the acetyl group is added to serotonin.
 |
| Exploring the Structure |
Exploring the Structure :
The KaiC clock protein is composed of six identical subunits that form a barrel-shaped structure. The protein is shown from side here, with two subunits removed to show the tunnel that runs through the middle. The phosphates that tick off the hours of each day are added to a serine and a threonine on each subunit. They are buried deep inside the tunnel, near the binding site for ATP, shown here in green. KaiA stimulates KaiC to add these phosphate groups to itself, and KaiB blocks the action of KaiA, allowing KaiC to remove these phosphates from itself. The speed of these reactions are all calibrated so that the whole process takes 24 hours to complete.


























 Online Movies
Online Movies
No comments:
Post a Comment